Student Contributor: Ms. Manisha De (BSc 3rd year Student, Department of Biochemistry, School of Life Science and Biotechnology)
Currently, Human existence is threatened by the newly emerged human coronaviruses (HCoVs) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing global pandemic disease outbreak (COVID-19). It has till now resulted in more than 1 lakh deaths worldwide and is losing its control as the number of days increases. A few years back in 2003 world was similarly threatened by a close SARS-CoV a close relative of SARS-CoV-2. That epidemic resulted in more than 8000 infections cases and 800 deaths which is much smaller in scale concerning COVID-19. Traditional public health measures used during SARS were successful and included active case detection, isolation of cases, contact tracing and quarantine of all contacts, social distancing, and community quarantine. Within 8 months these measures successfully able to eradicate SARS.
Its almost 4 months since the emergence of SARS-CoV-2, and there is hardly any remark of its stabilization. So, the question arises even there are many similarities between SARS-CoV and SARS-CoV-2 present whether the measures applicable during SARS-CoV can be effective during today’s pandemic?
Homology analysis: According to genome analyzing COVID-19(SARS-COV-2) has 89% nucleotide identity with batSARS-like-CoVZXC21 and 82% with that of human SARS-CoV. Like SARS-CoV and MERS-CoV, the SARS-COV-2 genome contains two flanking untranslated regions (UTRs) and a single long open reading frame(orf) encoding a polyprotein[1]. Further on comparing protein homology using NCBI Blastp the amino acid sequences of 28 proteins in SARS-CoV-2 were found homologous to SARS-CoV with an identity value≥65%, and query coverage≥95%[2].
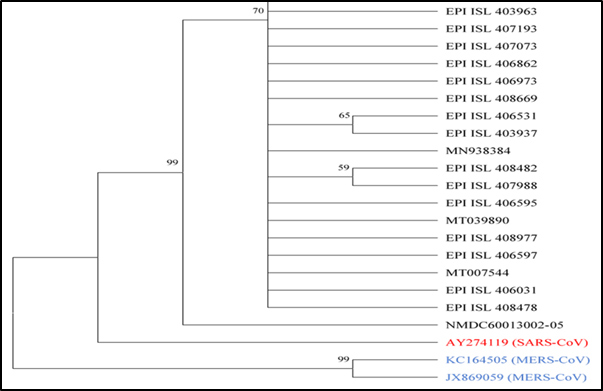
Phylogenetic Analysis:
With respect to evolutionary tree the distance of SARS-CoV (Accession no.- AY274119)
(Red coloured) is closer to SARS-CoV-2 (Accession no.-MN975262) strains than MERS-CoV (Accession no.- KC164505JX869059) (Blue coloured).
- Genomic comparison: There are six regions of difference (RD) in the genome sequence between SARS-CoV and SARS-CoV-2; RD1, RD2, and RD3 (448nt, 55nt, and 278nt, respectively) are partial coding sequences of the orf lab gene; RD4 and RD5 (315nt and 80nt, respectively) are partial coding sequences of the S gene; RD6 is 214nt in size and is part of the coding sequence of the orf7b and orf8 genes. These RDs may help in the development of new vaccines/drugs against SARS-CoV-2.
Proteomic comparison: Although proteins from SARS and SARS-CoV-2 were treated as homologous, two proteins (orf8 and orf10) in SARS-CoV-2 are not homologous to that in SARS-CoV. They differ in the amino acid sequence of orf8 in SARS-CoV-2 [3].Orf8 protein of SARS-CoV-2 does not contain a known functional domain or motif. Therefore, it will be clinically effective to analyze the biological function of these two specific proteins (orf8 and orf10) in SARS-CoV-2. Spike protein S2 in SARS-CoV-2 is highly conserved and shares 99% identity with those of the two bats SARS-like CoVs (bat-SL-CoVZXC21 and bat-SL-CoVZC45) and human SARS-CoV [3]. Thus, the broad-spectrum antiviral peptides against S2 have the potential to be effective treatment [4].
Transmission power: On another note, the transmissibility might be higher for COVID19 than for SARS. R₀ is a central concept in infectious disease epidemiology, indicating the risk of an infectious agent for its epidemic potential. A recent review (published in February 2020) found the average R₀ of COVID19 to be 3·28 and median R₀ to be 2·79, higher than that of SARS [5]. Despite massive containment efforts—it is certainly much faster than that reported for SARS between November 2002, and March 2003.
Infectious period: In contrast to COVID-19, identification was comparatively easier, and also isolation was effective for SARS as the peak viral shedding occurred after patients were quite ill with respiratory symptoms. Although mildly symptomatic patients have been reported for SARS, no known transmission occurred from these patients. Whereas, transmission during the early phases of illness was found to contribute to COVID-19 cases. Therefore, at the time of widespread community transmission is already evident for COVID19.
CFR (Case Fatality Ratio): Even if the CFR of COVID19 (possibly <2%) is far lower than that of SARS (10%), this is not reassuring, as a highly transmissible disease with low CFR will result in many more cases, and therefore also ultimately more deaths than SARS.[4]
Therefore, as there is no effective therapeutics or vaccines, the best way to deal with severe infections of CoVs is to control the source of infection, early diagnosis, reporting, isolation, supportive treatments, and timely publishing epidemic information to avoid unnecessary panic. For individuals, good personal hygiene, fitted masks, ventilation, and avoiding crowded places will help to prevent CoVs infection.
Reference:
- Chan, et al. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging microbes & infections, 9(1), 221-236.
- Xu, J et al. (2020). Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses, 12(2), 244.
- Chan, J et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes. Infect. 2020, 9, 221–236.
- Xia, S et al.A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019, 5, eaav4580
- Wilder-Smith, A et al. (2020). Can we contain the COVID-19 outbreak with the same measures as for SARS?. The Lancet Infectious Diseases.
Visited 2280 times, 3 Visits today



